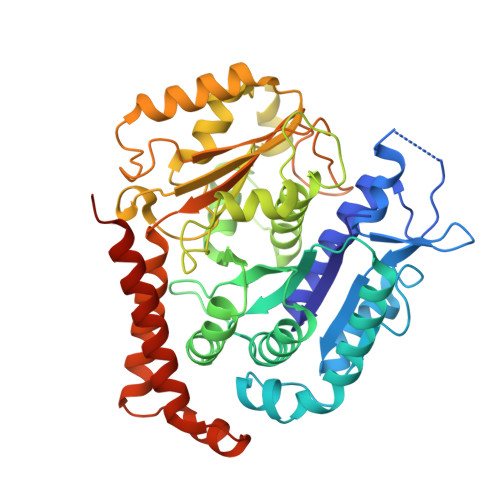
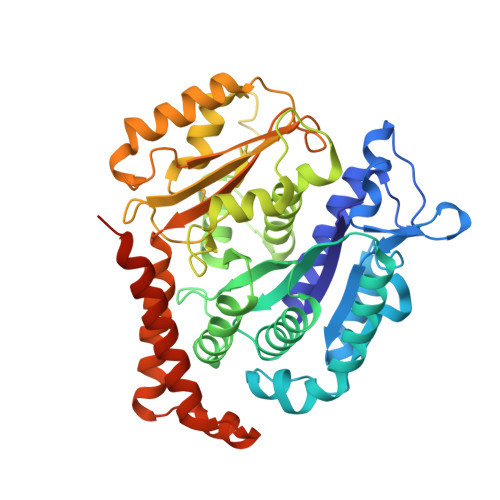
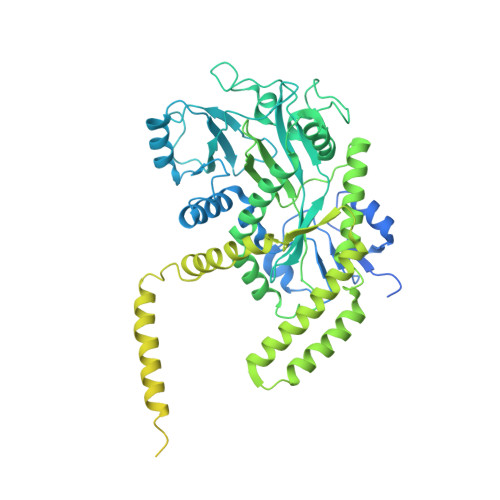
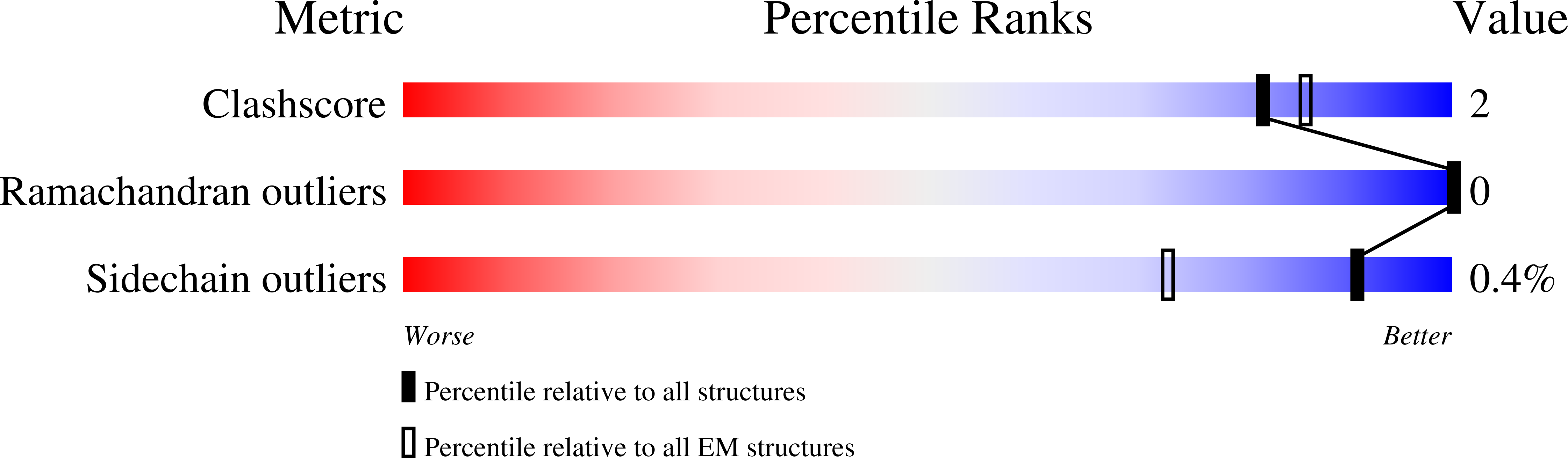Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Mahalingan, K.K., Grotjahn, D.A., Li, Y., Lander, G.C., Zehr, E.A., Roll-Mecak, A.(2024) Nat Chem Biol
- PubMed: 38658656
- DOI: https://doi.org/10.1038/s41589-024-01599-0
- Primary Citation of Related Structures:
8T42, 8U3Z - PubMed Abstract:
Microtubules have spatiotemporally complex posttranslational modification patterns. Tubulin tyrosine ligase-like (TTLL) enzymes introduce the most prevalent modifications on α-tubulin and β-tubulin. How TTLLs specialize for specific substrate recognition and ultimately modification-pattern generation is largely unknown. TTLL6, a glutamylase implicated in ciliopathies, preferentially modifies tubulin α-tails in microtubules. Cryo-electron microscopy, kinetic analysis and single-molecule biochemistry reveal an unprecedented quadrivalent recognition that ensures simultaneous readout of microtubule geometry and posttranslational modification status. By binding to a β-tubulin subunit, TTLL6 modifies the α-tail of the longitudinally adjacent tubulin dimer. Spanning two tubulin dimers along and across protofilaments (PFs) ensures fidelity of recognition of both the α-tail and the microtubule. Moreover, TTLL6 reads out and is stimulated by glutamylation of the β-tail of the laterally adjacent tubulin dimer, mediating crosstalk between α-tail and β-tail. This positive feedback loop can generate localized microtubule glutamylation patterns. Our work uncovers general principles that generate tubulin chemical and topographic complexity.
Organizational Affiliation:
Cell Biology and Biophysics Unit, Porter Neuroscience Research Center, National Institute of Neurological Disorders and Stroke, Bethesda, MD, USA.